ANOVA: One-way & Two-way (with Examples)
What is ANOVA?
Analysis of Variance (ANOVA) is a statistical method used to test whether the means of two or more groups are equal. ANOVA is an extension of the two-sample t-test to settings with more than two groups.
ANOVA focuses on how the total variability in a numeric outcome can be decomposed into: - Between-group variability (differences between group means), - Within-group variability (variability of observations inside the same group).
One-way ANOVA
A one-way ANOVA compares the means of a numeric variable across levels of a single categorical variable.
Example: a marketing department wants to know whether three teams have the same average sales performance.
Hypotheses
- \(H_0\): All group means are equal.
- \(H_1\): At least one group mean differs.
Assumptions (typical)
- Independent observations.
- Approximately normal residuals within each group.
- Equal variances across groups (homoscedasticity).
Example: one-way ANOVA with the poisons dataset
We use the poisons dataset:
time: survival timepoison: poison type (3 levels)treat: treatment type (3 levels)
Step 1) Import and prepare the data
library(dplyr)
library(readr)
path <- "raw_data/poisons.csv"
df <- read_csv(path, show_col_types = FALSE) |>
# robust: drop index column only if present
select(-any_of(c("X", "x"))) |>
mutate(
poison = factor(poison),
treat = factor(treat)
)
glimpse(df)## Rows: 48
## Columns: 4
## $ ...1 <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, …
## $ time <dbl> 0.31, 0.45, 0.46, 0.43, 0.36, 0.29, 0.40, 0.23, 0.22, 0.21, 0.1…
## $ poison <fct> 1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 1, 1, 1, 1, 2, 2, 2, 2, 3, …
## $ treat <fct> A, A, A, A, A, A, A, A, A, A, A, A, B, B, B, B, B, B, B, B, B, …Step 2) Summary statistics by group
df |>
dplyr::group_by(poison) |>
dplyr::summarize(
n = n(),
mean_time = mean(time, na.rm = TRUE),
sd_time = sd(time, na.rm = TRUE),
.groups = "drop"
)| poison | n | mean_time | sd_time |
|---|---|---|---|
| 1 | 16 | 0.617500 | 0.2094278 |
| 2 | 16 | 0.544375 | 0.2893664 |
| 3 | 16 | 0.276250 | 0.0622763 |
Step 3) Visual check (boxplot + jitter)
library(ggplot2)
ggplot(df, aes(x = poison, y = time, fill = poison)) +
geom_boxplot(alpha = 0.7, width = 0.65, outlier.shape = NA) +
geom_jitter(width = 0.15, alpha = 0.6, size = 1.6, color = "steelblue") +
theme_classic() +
guides(fill = "none")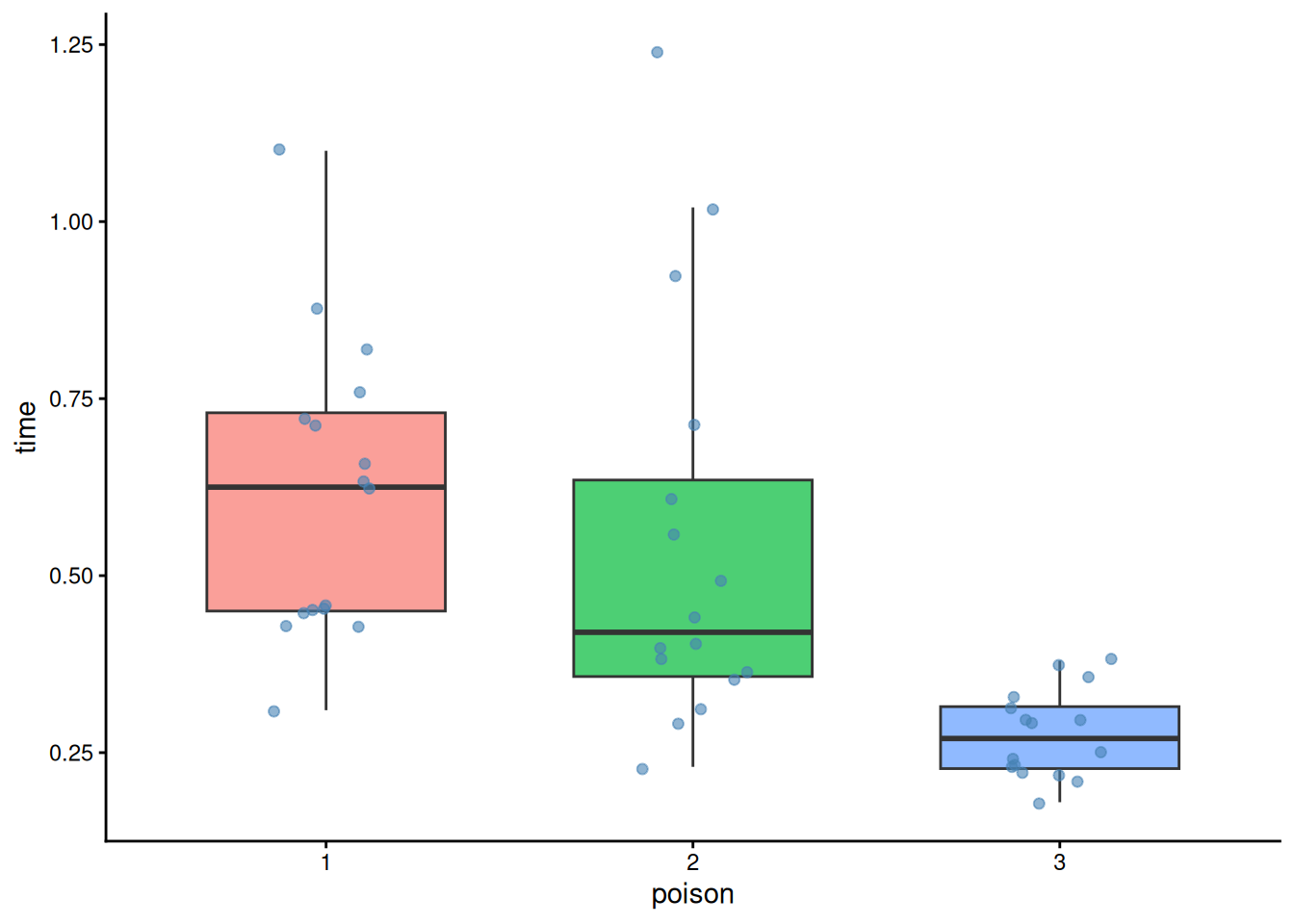
Step 4) Fit the one-way ANOVA
## Df Sum Sq Mean Sq F value Pr(>F)
## poison 2 1.033 0.5165 11.79 7.66e-05 ***
## Residuals 45 1.972 0.0438
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Step 5) Post-hoc comparisons (Tukey HSD)
The ANOVA tells you whether any difference exists, but not which pairs differ. Tukey’s HSD provides pairwise comparisons with multiple-testing adjustment.
## Tukey multiple comparisons of means
## 95% family-wise confidence level
##
## Fit: aov(formula = time ~ poison, data = df)
##
## $poison
## diff lwr upr p adj
## 2-1 -0.073125 -0.2525046 0.10625464 0.5881654
## 3-1 -0.341250 -0.5206296 -0.16187036 0.0000971
## 3-2 -0.268125 -0.4475046 -0.08874536 0.0020924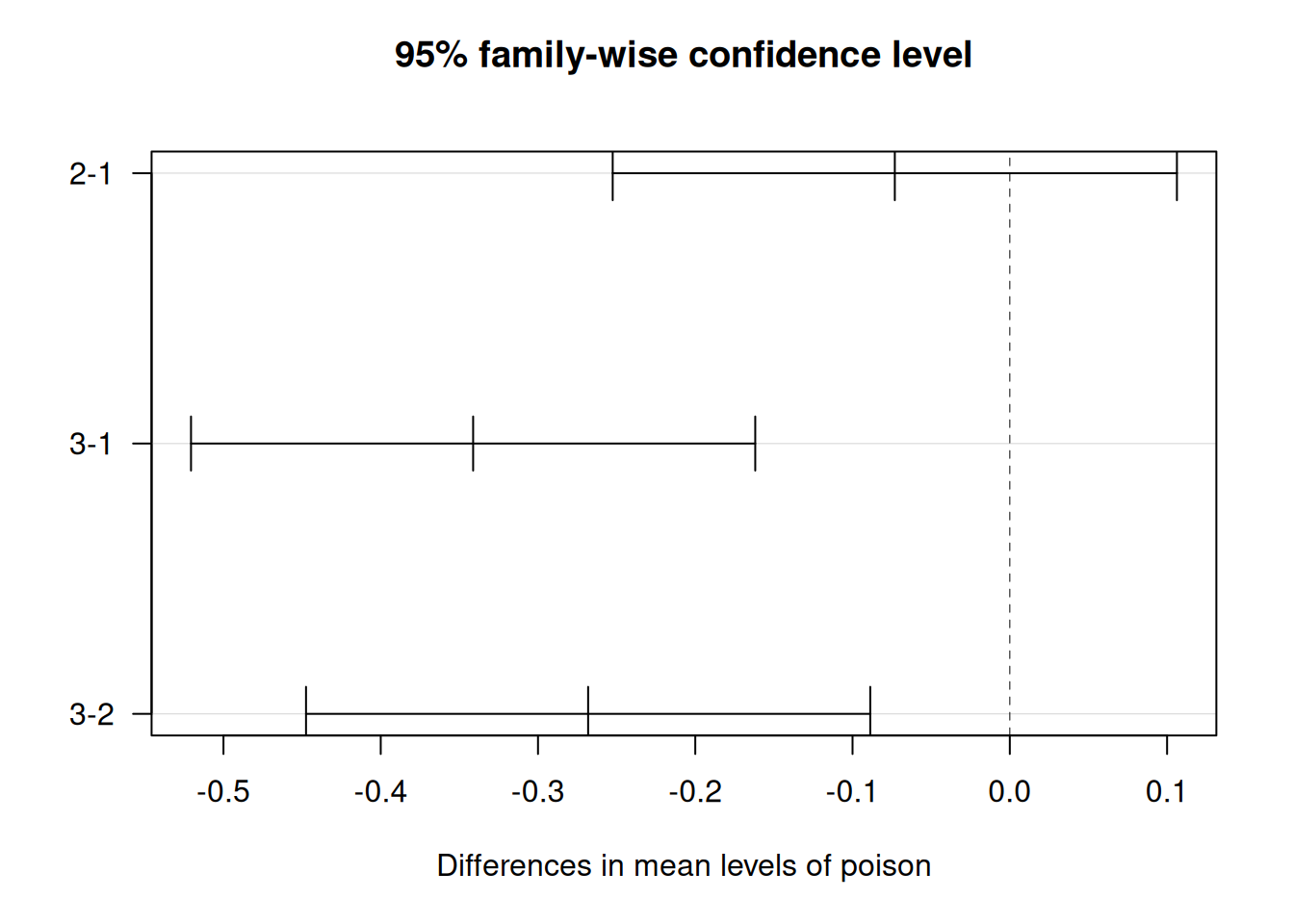
Two-way ANOVA
A two-way ANOVA includes two categorical predictors.
You typically consider: - Main effects: poison and
treat - Interaction: whether the effect of treatment
depends on poison type
Additive model (main effects only)
## Df Sum Sq Mean Sq F value Pr(>F)
## poison 2 1.0330 0.5165 20.64 5.7e-07 ***
## treat 3 0.9212 0.3071 12.27 6.7e-06 ***
## Residuals 42 1.0509 0.0250
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Interaction model (recommended to check)
## Df Sum Sq Mean Sq F value Pr(>F)
## poison 2 1.0330 0.5165 23.222 3.33e-07 ***
## treat 3 0.9212 0.3071 13.806 3.78e-06 ***
## poison:treat 6 0.2501 0.0417 1.874 0.112
## Residuals 36 0.8007 0.0222
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1If the interaction term is significant, interpret the interaction first (effects are not constant across levels of the other factor).
Summary
| Test | Code |
|---|---|
| One-way ANOVA | aov(time ~ poison, data = df) |
| Post-hoc (pairwise) | TukeyHSD(anova_one_way) |
| Two-way ANOVA (additive) | aov(time ~ poison + treat, data = df) |
| Two-way ANOVA (interaction) | aov(time ~ poison * treat, data = df) |
A work by Gianluca Sottile
gianluca.sottile@unipa.it