Gradient Boosting in R (XGBoost) — Titanic Classification
Gradient Boosting (XGBoost) on Titanic
Gradient boosting builds trees sequentially: each new tree focuses on correcting the mistakes of the previous ensemble. XGBoost is a highly optimized implementation of gradient boosting with practical features like regularization and early stopping.
In this lesson we will: - Import Titanic data from a URL - Clean and encode features for XGBoost - Create train/test split - Train XGBoost for binary classification - Evaluate accuracy + confusion matrix - Use cross-validation with early stopping - Inspect feature importance
Step 1: Import the data
library(dplyr)
path <- "raw_data/titanic_data.csv"
titanic <- read.csv(path, stringsAsFactors = FALSE)
dim(titanic)## [1] 1309 13| x | pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | home.dest |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | Allen, Miss. Elisabeth Walton | female | 29 | 0 | 0 | 24160 | 211.3375 | B5 | S | St Louis, MO |
| 2 | 1 | 1 | Allison, Master. Hudson Trevor | male | 0.9167 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
| 3 | 1 | 0 | Allison, Miss. Helen Loraine | female | 2 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
Step 2: Clean and prepare data
We follow a simple approach similar to the Decision Tree lesson: - Drop columns that are not useful or require heavy feature engineering (IDs, names, ticket, cabin, etc.) - Convert categorical variables to factors - Remove missing values (for simplicity)
titanic_clean <- titanic |>
select(-c(home.dest, cabin, name, x, ticket)) |>
filter(embarked != "?") |>
mutate(
pclass = factor(
pclass,
levels = c(1, 2, 3),
labels = c("Upper", "Middle", "Lower")
),
survived = factor(survived, levels = c(0, 1), labels = c("No", "Yes")),
sex = factor(sex),
embarked = factor(embarked),
age = as.numeric(age),
fare = as.numeric(fare)
) |>
na.omit()
glimpse(titanic_clean)## Rows: 1,043
## Columns: 8
## $ pclass <fct> Upper, Upper, Upper, Upper, Upper, Upper, Upper, Upper, Upper…
## $ survived <fct> Yes, Yes, No, No, No, Yes, Yes, No, Yes, No, No, Yes, Yes, Ye…
## $ sex <fct> female, male, female, male, female, male, female, male, femal…
## $ age <dbl> 29.0000, 0.9167, 2.0000, 30.0000, 25.0000, 48.0000, 63.0000, …
## $ sibsp <int> 0, 1, 1, 1, 1, 0, 1, 0, 2, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1…
## $ parch <int> 0, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1…
## $ fare <dbl> 211.3375, 151.5500, 151.5500, 151.5500, 151.5500, 26.5500, 77…
## $ embarked <fct> S, S, S, S, S, S, S, S, S, C, C, C, C, S, S, C, C, C, C, S, S…Why we need encoding
XGBoost expects a numeric matrix. A common and reliable approach is
one-hot encoding via model.matrix().
# Create design matrix (one-hot encoding for factors)
X_all <- model.matrix(survived ~ . - 1, data = titanic_clean)
# Label as 0/1 (XGBoost wants numeric labels for binary logistic)
y_all <- ifelse(titanic_clean$survived == "Yes", 1, 0)
dim(X_all)## [1] 1043 10## y_all
## 0 1
## 618 425Step 3: Train/test split
set.seed(123)
n <- nrow(X_all)
idx_train <- sample.int(n, size = floor(0.8 * n))
X_train <- X_all[idx_train, , drop = FALSE]
y_train <- y_all[idx_train]
X_test <- X_all[-idx_train, , drop = FALSE]
y_test <- y_all[-idx_train]
c(n_train = nrow(X_train), n_test = nrow(X_test))## n_train n_test
## 834 209Step 4: Train a baseline XGBoost model
We use binary logistic objective: - output is a probability in [0, 1] - we then choose a threshold (e.g., 0.5) for class prediction
library(xgboost)
dtrain <- xgb.DMatrix(data = X_train, label = y_train)
dtest <- xgb.DMatrix(data = X_test, label = y_test)
params <- list(
objective = "binary:logistic",
eval_metric = "logloss",
max_depth = 3,
eta = 0.1,
subsample = 0.8,
colsample_bytree = 0.8
)
set.seed(123)
xgb_fit <- xgb.train(
params = params,
data = dtrain,
nrounds = 200,
watchlist = list(train = dtrain, test = dtest),
verbose = 0
)
xgb_fit## ##### xgb.Booster
## call:
## xgb.train(params = params, data = dtrain, nrounds = 200, verbose = 0,
## watchlist = list(train = dtrain, test = dtest))
## # of features: 10
## # of rounds: 200
## callbacks:
## evaluation_log
## evaluation_log:
## iter train_logloss test_logloss
## <num> <num> <num>
## 1 0.6614907 0.6512885
## 2 0.6479067 0.6352419
## --- --- ---
## 199 0.2870097 0.4270068
## 200 0.2865113 0.4279921Step 5: Predictions and evaluation
pred_prob <- predict(xgb_fit, newdata = dtest)
pred_class <- ifelse(pred_prob >= 0.5, 1, 0)
conf_mat <- table(actual = y_test, predicted = pred_class)
conf_mat## predicted
## actual 0 1
## 0 109 19
## 1 17 64## [1] 0.8277512Step 6: Cross-validation + early stopping (choose nrounds)
Early stopping finds the number of boosting rounds that minimizes the validation loss and stops when there is no improvement for some rounds.
set.seed(123)
cv <- xgb.cv(
params = params,
data = dtrain,
nrounds = 1000,
nfold = 5,
early_stopping_rounds = 25,
verbose = 0
)
cv$early_stop$best_iteration## [1] 84Train a final model using the best number of rounds:
best_nrounds <- cv$early_stop$best_iteration
set.seed(123)
xgb_final <- xgb.train(
params = params,
data = dtrain,
nrounds = best_nrounds,
watchlist = list(train = dtrain, test = dtest),
verbose = 0
)
pred_prob2 <- predict(xgb_final, newdata = dtest)
pred_class2 <- ifelse(pred_prob2 >= 0.5, 1, 0)
conf_mat2 <- table(actual = y_test, predicted = pred_class2)
conf_mat2## predicted
## actual 0 1
## 0 114 14
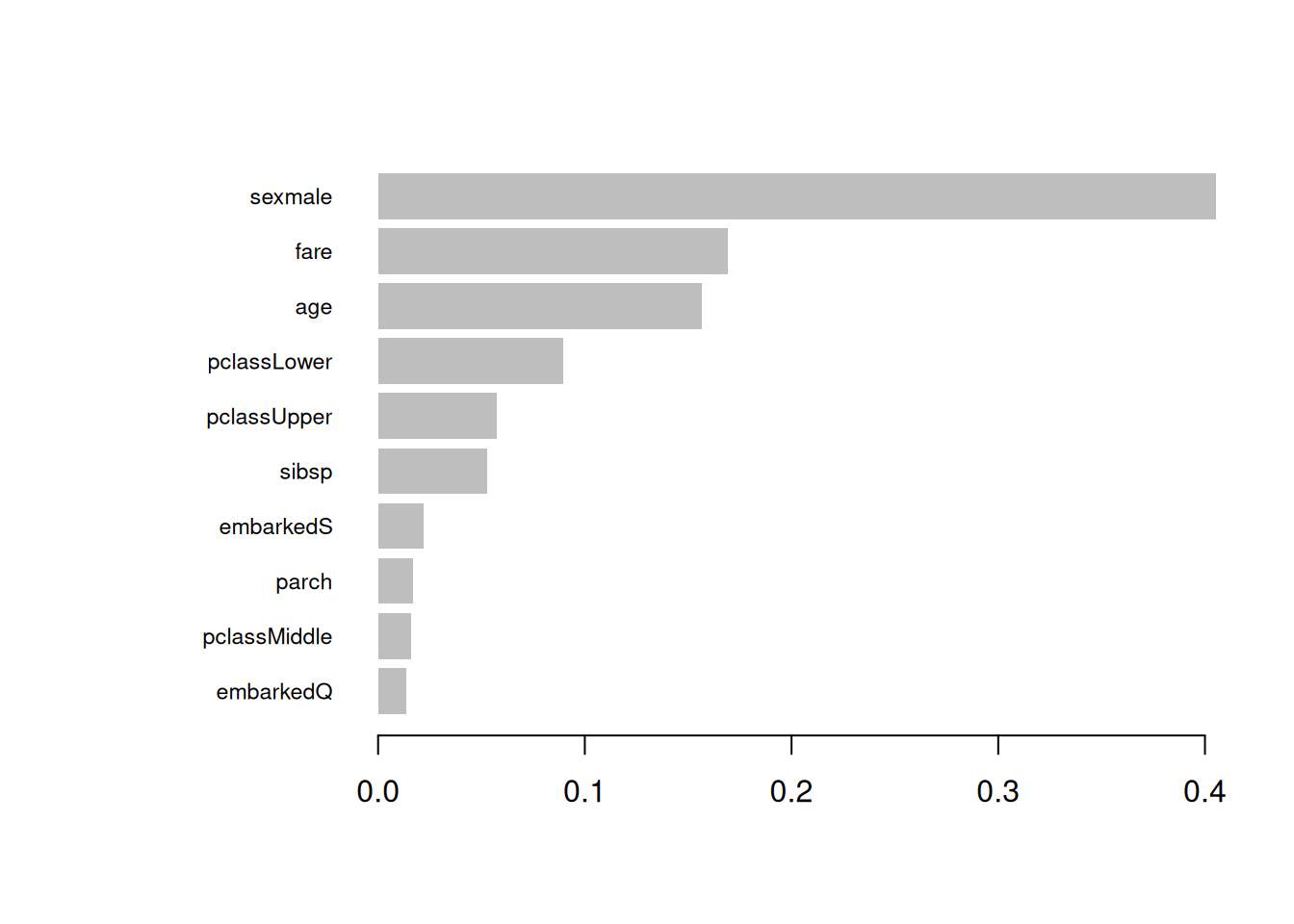
## 1 18 63## [1] 0.84689Step 7: Feature importance
| Feature | Gain | Cover | Frequency |
|---|---|---|---|
| sexmale | 0.4055162 | 0.1406440 | 0.0855513 |
| fare | 0.1693183 | 0.2846434 | 0.3231939 |
| age | 0.1564935 | 0.2774125 | 0.2927757 |
| pclassLower | 0.0896165 | 0.0420888 | 0.0494297 |
| pclassUpper | 0.0576380 | 0.0555959 | 0.0399240 |
| sibsp | 0.0527441 | 0.0520967 | 0.0874525 |
| embarkedS | 0.0222473 | 0.0572843 | 0.0456274 |
| parch | 0.0168627 | 0.0287922 | 0.0247148 |
| pclassMiddle | 0.0161098 | 0.0223554 | 0.0209125 |
| embarkedQ | 0.0134537 | 0.0390868 | 0.0304183 |
(Optional) Step 8: Simple tuning grid
This is a small grid search to illustrate the idea (not exhaustive).
grid <- expand.grid(
max_depth = c(2, 3, 4),
eta = c(0.05, 0.1),
subsample = c(0.8, 1.0),
colsample_bytree = c(0.8, 1.0)
)
grid$best_iter <- NA_integer_
grid$best_auc <- NA_real_
set.seed(123)
for (i in seq_len(nrow(grid))) {
params_i <- list(
objective = "binary:logistic",
eval_metric = "auc",
max_depth = grid$max_depth[i],
eta = grid$eta[i],
subsample = grid$subsample[i],
colsample_bytree = grid$colsample_bytree[i]
)
cv_i <- xgb.cv(
params = params_i,
data = dtrain,
nrounds = 800,
nfold = 5,
early_stopping_rounds = 25,
verbose = 0
)
grid$best_iter[i] <- cv_i$early_stop$best_iteration
grid$best_auc[i] <- cv_i$evaluation_log$train_auc_mean[cv_i$early_stop$best_iteration]
}
grid <- grid[order(grid$best_auc, decreasing = TRUE), ]
head(grid, 10)| max_depth | eta | subsample | colsample_bytree | best_iter | best_auc | |
|---|---|---|---|---|---|---|
| 18 | 4 | 0.10 | 0.8 | 1.0 | 69 | 0.9497712 |
| 6 | 4 | 0.10 | 0.8 | 0.8 | 40 | 0.9273228 |
| 16 | 2 | 0.10 | 0.8 | 1.0 | 139 | 0.9179912 |
| 9 | 4 | 0.05 | 1.0 | 0.8 | 54 | 0.9171799 |
| 12 | 4 | 0.10 | 1.0 | 0.8 | 23 | 0.9094559 |
| 11 | 3 | 0.10 | 1.0 | 0.8 | 45 | 0.9085839 |
| 24 | 4 | 0.10 | 1.0 | 1.0 | 19 | 0.9058019 |
| 14 | 3 | 0.05 | 0.8 | 1.0 | 71 | 0.9055522 |
| 15 | 4 | 0.05 | 0.8 | 1.0 | 32 | 0.9045788 |
| 4 | 2 | 0.10 | 0.8 | 0.8 | 97 | 0.9042485 |
Summary
You learned how to: - clean and encode Titanic data for XGBoost - train a gradient boosting model for binary classification - evaluate with a confusion matrix and accuracy - use cross-validation + early stopping to pick the number of trees - inspect feature importance
A work by Gianluca Sottile
gianluca.sottile@unipa.it