Decision Trees in R (rpart): Classification Example with Titanic
Decision trees in R
A decision tree is a supervised learning model that can be used for: - classification (predicting classes) - regression (predicting numeric outcomes)
In this lesson we build a simple classification tree
in R using rpart, then evaluate and tune it.
Step 1: Import the data
We will use a Titanic dataset hosted online.
path <- "raw_data/titanic_data.csv"
titanic <- read.csv(path, stringsAsFactors = FALSE)
dim(titanic)## [1] 1309 13| x | pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | home.dest |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | Allen, Miss. Elisabeth Walton | female | 29 | 0 | 0 | 24160 | 211.3375 | B5 | S | St Louis, MO |
| 2 | 1 | 1 | Allison, Master. Hudson Trevor | male | 0.9167 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
| 3 | 1 | 0 | Allison, Miss. Helen Loraine | female | 2 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
Step 2: Clean the dataset
Typical cleaning steps for this dataset: - drop high-cardinality or irrelevant columns - create factors for categorical variables - handle missing values
library(dplyr)
titanic_clean <- titanic |>
select(-c(home.dest, cabin, name, x, ticket)) |>
mutate(
pclass = factor(
pclass,
levels = c(1, 2, 3),
labels = c("Upper", "Middle", "Lower")
),
survived = factor(
survived,
levels = c(0, 1),
labels = c("No", "Yes")
),
sex = factor(sex),
embarked = factor(embarked),
age = as.numeric(age),
fare = as.numeric(fare)
) |>
na.omit()
glimpse(titanic_clean)## Rows: 1,045
## Columns: 8
## $ pclass <fct> Upper, Lower, Lower, Middle, Lower, Middle, Lower, Lower, Upp…
## $ survived <fct> Yes, No, No, No, No, No, No, No, Yes, No, Yes, No, No, Yes, N…
## $ sex <fct> male, male, male, male, female, female, male, male, female, m…
## $ age <dbl> 36.0, 42.0, 18.5, 44.0, 19.0, 26.0, 23.0, 28.5, 64.0, 36.5, 4…
## $ sibsp <int> 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0…
## $ parch <int> 2, 0, 0, 0, 0, 1, 0, 0, 2, 2, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0…
## $ fare <dbl> 120.0000, 8.6625, 7.2292, 13.0000, 16.1000, 26.0000, 7.8542, …
## $ embarked <fct> S, S, C, S, S, S, S, S, C, S, S, S, S, S, S, S, S, S, Q, S, S…Step 3: Train/test split
A simple and reproducible split that returns both datasets.
split_data <- function(df, prop = 0.8, seed = NULL) {
stopifnot(is.data.frame(df))
stopifnot(prop > 0 && prop < 1)
if (!is.null(seed)) set.seed(seed)
n <- nrow(df)
n_train <- floor(prop * n)
idx_train <- sample.int(n, size = n_train)
list(
train = df[idx_train, , drop = FALSE],
test = df[-idx_train, , drop = FALSE]
)
}
spl <- split_data(titanic_clean, prop = 0.8, seed = 123)
train <- spl$train
test <- spl$test
dim(train)## [1] 836 8## [1] 209 8##
## No Yes
## 0.5933014 0.4066986##
## No Yes
## 0.5837321 0.4162679Step 4: Fit a classification tree (rpart)
## n= 836
##
## node), split, n, loss, yval, (yprob)
## * denotes terminal node
##
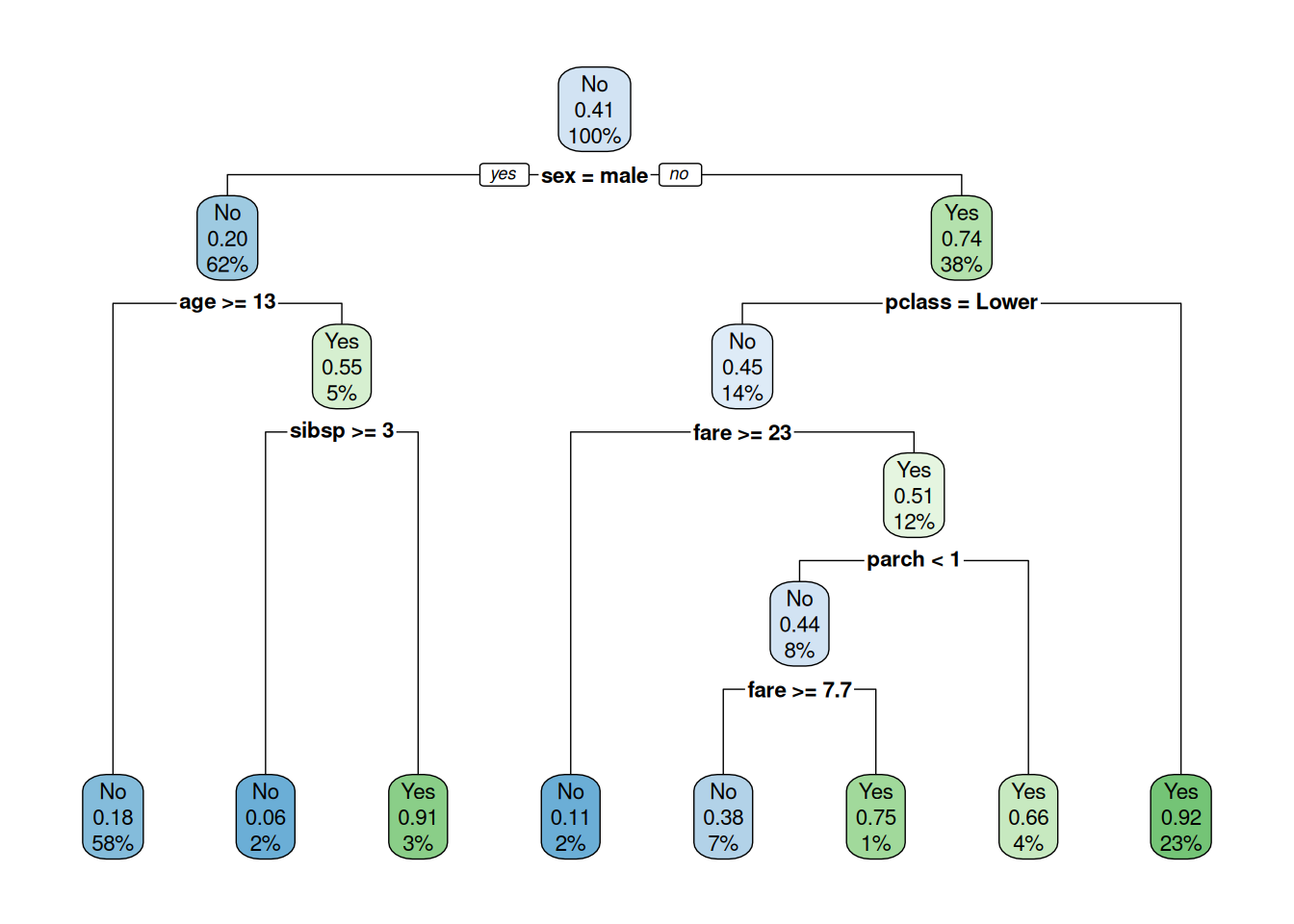
## 1) root 836 340 No (0.59330144 0.40669856)
## 2) sex=male 521 106 No (0.79654511 0.20345489)
## 4) age>=12.5 483 85 No (0.82401656 0.17598344) *
## 5) age< 12.5 38 17 Yes (0.44736842 0.55263158)
## 10) sibsp>=2.5 16 1 No (0.93750000 0.06250000) *
## 11) sibsp< 2.5 22 2 Yes (0.09090909 0.90909091) *
## 3) sex=female 315 81 Yes (0.25714286 0.74285714)
## 6) pclass=Lower 121 55 No (0.54545455 0.45454545)
## 12) fare>=23.0875 18 2 No (0.88888889 0.11111111) *
## 13) fare< 23.0875 103 50 Yes (0.48543689 0.51456311)
## 26) parch< 0.5 68 30 No (0.55882353 0.44117647)
## 52) fare>=7.72915 56 21 No (0.62500000 0.37500000) *
## 53) fare< 7.72915 12 3 Yes (0.25000000 0.75000000) *
## 27) parch>=0.5 35 12 Yes (0.34285714 0.65714286) *
## 7) pclass=Upper,Middle 194 15 Yes (0.07731959 0.92268041) *Step 5: Predict on the test set
## 57 920 653 348 354 54
## No No No No Yes No
## Levels: No YesStep 6: Evaluate performance
A quick evaluation uses a confusion matrix and accuracy.
## predicted
## actual No Yes
## No 113 9
## Yes 31 56## [1] 0.8086124Tip: for imbalanced datasets, also consider metrics like sensitivity/recall, specificity, precision, and F1-score.
Step 7: Tune hyperparameters (simple grid search)
Below is a minimal tuning loop over a few
rpart.control() parameters. This is intentionally simple
for learning; in real projects you would likely use
cross-validation.
accuracy_tree <- function(fit, test_df) {
pred <- predict(fit, newdata = test_df, type = "class")
cm <- table(test_df$survived, pred)
sum(diag(cm)) / sum(cm)
}
grid <- expand.grid(
maxdepth = c(2, 3, 4, 5),
minsplit = c(2, 5, 10, 20),
stringsAsFactors = FALSE
)
results <- grid
results$accuracy <- NA_real_
for (i in seq_len(nrow(grid))) {
ctrl <- rpart.control(
maxdepth = grid$maxdepth[i],
minsplit = grid$minsplit[i],
cp = 0
)
fit_i <- rpart(
survived ~ .,
data = train,
method = "class",
control = ctrl
)
results$accuracy[i] <- accuracy_tree(fit_i, test)
}
results <- results[order(-results$accuracy), ]
head(results, 10)| maxdepth | minsplit | accuracy | |
|---|---|---|---|
| 2 | 3 | 2 | 0.8181818 |
| 6 | 3 | 5 | 0.8181818 |
| 10 | 3 | 10 | 0.8181818 |
| 14 | 3 | 20 | 0.8181818 |
| 4 | 5 | 2 | 0.8038278 |
| 8 | 5 | 5 | 0.8038278 |
| 16 | 5 | 20 | 0.7990431 |
| 15 | 4 | 20 | 0.7894737 |
| 3 | 4 | 2 | 0.7846890 |
| 7 | 4 | 5 | 0.7846890 |
Summary
In this lesson you learned how to: - import and clean data - split
train/test properly (with shuffling + reproducibility) - fit a
classification tree with rpart - visualize the model with
rpart.plot - evaluate accuracy and run a simple tuning
loop
A work by Gianluca Sottile
gianluca.sottile@unipa.it