Random Forest in R (caret + randomForest) — Titanic Classification
Random Forest on Titanic
A Random Forest is an ensemble of many decision trees that can be used for classification and regression. In this lesson we follow the same workflow as the Decision Tree / ML lessons: import → clean → split → fit → evaluate → tune → interpret.
In this lesson we will: - Import Titanic data from a local CSV -
Clean the dataset (drop columns + fix types) - Create train/test split -
Train a Random Forest classifier with cross-validation - Evaluate
accuracy + confusion matrix - Tune mtry (number of
candidate predictors per split) - Inspect feature importance
Step 1: Import the data
library(dplyr)
path <- "raw_data/titanic_data.csv"
titanic <- read.csv(path, stringsAsFactors = FALSE)
dim(titanic)## [1] 1309 13| x | pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | home.dest |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | Allen, Miss. Elisabeth Walton | female | 29 | 0 | 0 | 24160 | 211.3375 | B5 | S | St Louis, MO |
| 2 | 1 | 1 | Allison, Master. Hudson Trevor | male | 0.9167 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
| 3 | 1 | 0 | Allison, Miss. Helen Loraine | female | 2 | 1 | 2 | 113781 | 151.55 | C22 C26 | S | Montreal, PQ / Chesterville, ON |
Step 2: Clean and prepare data
Random Forest (via randomForest) can handle factors
directly, so we typically do not need one-hot encoding
like XGBoost.
titanic_clean <- titanic |>
# drop columns that are not useful or require heavy feature engineering
select(-any_of(c("home.dest", "cabin", "name", "X", "x", "ticket"))) |>
# remove invalid entries sometimes used in this dataset
filter(embarked != "?") |>
mutate(
pclass = factor(
pclass,
levels = c(1, 2, 3),
labels = c("Upper", "Middle", "Lower")
),
survived = factor(survived, levels = c(0, 1), labels = c("No", "Yes")),
sex = factor(sex),
embarked = factor(embarked),
age = as.numeric(age),
fare = as.numeric(fare),
sibsp = as.numeric(sibsp),
parch = as.numeric(parch)
) |>
na.omit()
glimpse(titanic_clean)## Rows: 1,043
## Columns: 8
## $ pclass <fct> Upper, Upper, Upper, Upper, Upper, Upper, Upper, Upper, Upper…
## $ survived <fct> Yes, Yes, No, No, No, Yes, Yes, No, Yes, No, No, Yes, Yes, Ye…
## $ sex <fct> female, male, female, male, female, male, female, male, femal…
## $ age <dbl> 29.0000, 0.9167, 2.0000, 30.0000, 25.0000, 48.0000, 63.0000, …
## $ sibsp <dbl> 0, 1, 1, 1, 1, 0, 1, 0, 2, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1…
## $ parch <dbl> 0, 2, 2, 2, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1…
## $ fare <dbl> 211.3375, 151.5500, 151.5500, 151.5500, 151.5500, 26.5500, 77…
## $ embarked <fct> S, S, S, S, S, S, S, S, S, C, C, C, C, S, S, C, C, C, C, S, S…Step 3: Train/test split
set.seed(123)
n <- nrow(titanic_clean)
idx_train <- sample.int(n, size = floor(0.8 * n))
train_df <- titanic_clean[idx_train, ]
test_df <- titanic_clean[-idx_train, ]
c(n_train = nrow(train_df), n_test = nrow(test_df))## n_train n_test
## 834 209##
## No Yes
## 0.58753 0.41247##
## No Yes
## 0.6124402 0.3875598Step 4: Train a baseline Random Forest (cross-validation)
We will use caret::train() with
method = "rf", which fits a Random Forest and tunes
mtry using resampling. In caret, the tuning
grid for method="rf" is expected to include
only mtry (while ntree is
passed as a regular argument, not as a tuning parameter).
# install.packages(c("caret", "randomForest", "e1071"))
library(caret)
library(randomForest)
library(e1071)
set.seed(123)
ctrl <- trainControl(
method = "cv",
number = 5,
classProbs = TRUE
)
rf_cv <- train(
survived ~ .,
data = train_df,
method = "rf",
metric = "Accuracy",
trControl = ctrl,
tuneLength = 6, # caret tries multiple mtry values
ntree = 500,
importance = TRUE
)
rf_cv## Random Forest
##
## 834 samples
## 7 predictor
## 2 classes: 'No', 'Yes'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 668, 667, 667, 667, 667
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 2 0.7913643 0.5509943
## 3 0.7829738 0.5364574
## 4 0.7841931 0.5425450
## 6 0.7745906 0.5273374
## 7 0.7721809 0.5220312
## 9 0.7650025 0.5076265
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 2.| mtry |
|---|
| 2 |
Step 5: Predictions
## [1] Yes Yes Yes Yes Yes No
## Levels: No YesStep 6: Evaluation (confusion matrix + accuracy)
## Confusion Matrix and Statistics
##
## Reference
## Prediction No Yes
## No 117 21
## Yes 11 60
##
## Accuracy : 0.8469
## 95% CI : (0.7908, 0.8929)
## No Information Rate : 0.6124
## P-Value [Acc > NIR] : 1.041e-13
##
## Kappa : 0.67
##
## Mcnemar's Test P-Value : 0.1116
##
## Sensitivity : 0.9141
## Specificity : 0.7407
## Pos Pred Value : 0.8478
## Neg Pred Value : 0.8451
## Prevalence : 0.6124
## Detection Rate : 0.5598
## Detection Prevalence : 0.6603
## Balanced Accuracy : 0.8274
##
## 'Positive' Class : No
## Step 7: Feature importance
The randomForest model can report variable importance
(e.g., MeanDecreaseAccuracy and MeanDecreaseGini in classification when
importance is enabled).
## rf variable importance
##
## Importance
## sexmale 100.000
## pclassLower 25.815
## age 19.443
## fare 15.149
## embarkedS 6.385
## pclassMiddle 2.383
## parch 2.207
## sibsp 1.491
## embarkedQ 0.000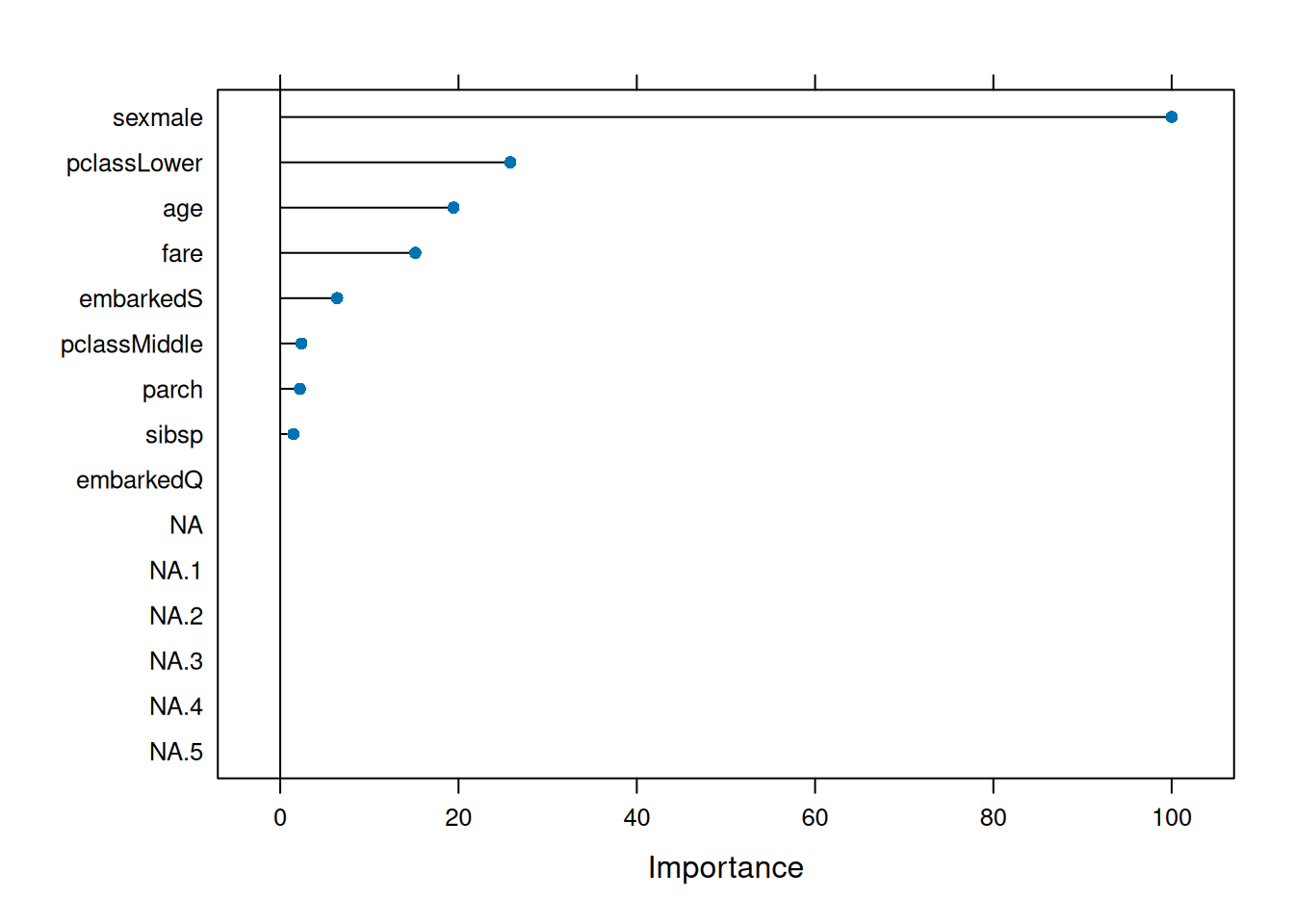
# randomForest-style importance (from the final fitted model)
rf_final <- rf_cv$finalModel
imp_rf <- importance(rf_final)
head(imp_rf[order(imp_rf[, "MeanDecreaseGini"], decreasing = TRUE), , drop = FALSE], 10)## No Yes MeanDecreaseAccuracy MeanDecreaseGini
## sexmale 47.845292 55.834089 56.960665 86.791194
## fare 10.735517 12.988644 19.656992 42.849169
## age 18.719531 9.050861 22.038897 34.603448
## pclassLower 16.871365 16.902909 24.517388 19.365896
## parch 10.234109 1.294826 10.483391 10.853481
## sibsp 12.498860 -1.645138 10.465300 10.546136
## embarkedS 14.398329 1.066856 12.599433 8.046899
## pclassMiddle 9.011597 2.682556 10.164223 5.348113
## embarkedQ 11.146658 -1.697619 9.293445 2.839485(Optional) Step 8: Manual tuning grid for mtry
Here is a more explicit tuning grid for mtry; note that
the grid must have the column mtry only.
p <- ncol(train_df) - 1 # predictors count (excluding survived)
grid_mtry <- expand.grid(mtry = unique(pmax(1, round(c(1, sqrt(p), p/3, p/2)))))
set.seed(123)
rf_tuned <- train(
survived ~ .,
data = train_df,
method = "rf",
metric = "Accuracy",
trControl = ctrl,
tuneGrid = grid_mtry,
ntree = 500,
importance = TRUE
)
rf_tuned## Random Forest
##
## 834 samples
## 7 predictor
## 2 classes: 'No', 'Yes'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 668, 667, 667, 667, 667
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 1 0.7649953 0.4788094
## 2 0.7913715 0.5497244
## 3 0.7841714 0.5393207
## 4 0.7806002 0.5352287
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 2.| mtry | |
|---|---|
| 2 | 2 |
Evaluate the tuned model on the test set:
## Confusion Matrix and Statistics
##
## Reference
## Prediction No Yes
## No 117 22
## Yes 11 59
##
## Accuracy : 0.8421
## 95% CI : (0.7855, 0.8888)
## No Information Rate : 0.6124
## P-Value [Acc > NIR] : 3.586e-13
##
## Kappa : 0.6589
##
## Mcnemar's Test P-Value : 0.08172
##
## Sensitivity : 0.9141
## Specificity : 0.7284
## Pos Pred Value : 0.8417
## Neg Pred Value : 0.8429
## Prevalence : 0.6124
## Detection Rate : 0.5598
## Detection Prevalence : 0.6651
## Balanced Accuracy : 0.8212
##
## 'Positive' Class : No
## Summary
You learned how to: - clean Titanic data for Random Forest without
one-hot encoding - train a Random Forest classifier with
cross-validation using caret - tune mtry and
keep ntree fixed during training - evaluate performance
using a confusion matrix - inspect variable importance from the fitted
forest
A work by Gianluca Sottile
gianluca.sottile@unipa.it